Ubiquitin and Signal Transduction
Background
The ubiquitin pathway regulates many biological processes and determines how cells respond to growth factors, stress and genetic damage, controlling nearly every facet of a cell’s life and death. The covalent attachment of ubiquitin to proteins is not simply a sign of destruction but it can be a regulatory signal that alters localization, activity, and ultimate fate of the target proteins.
Ubiquitination comes in multiple flavors, ranging from a simple monoubiquitination to polymeric ubiquitin chains with complex topologies. The existence of a vast and dynamic array of Ub polymers raises the central question of how specificity is achieved during conjugation, recognition, and signal transduction. Answer to this issue is a main object of our studies.
We study one of the most intriguing features of the ubiquitin, not directly related to protein degradation. Work by us and others in the last ten years led to the definition of ubiquitin as a signaling molecule, critical in the regulation of receptor tyrosine kinase pathways, represented in our lab by a model system of the epidermal growth factor receptor (EGFR) signaling pathway.
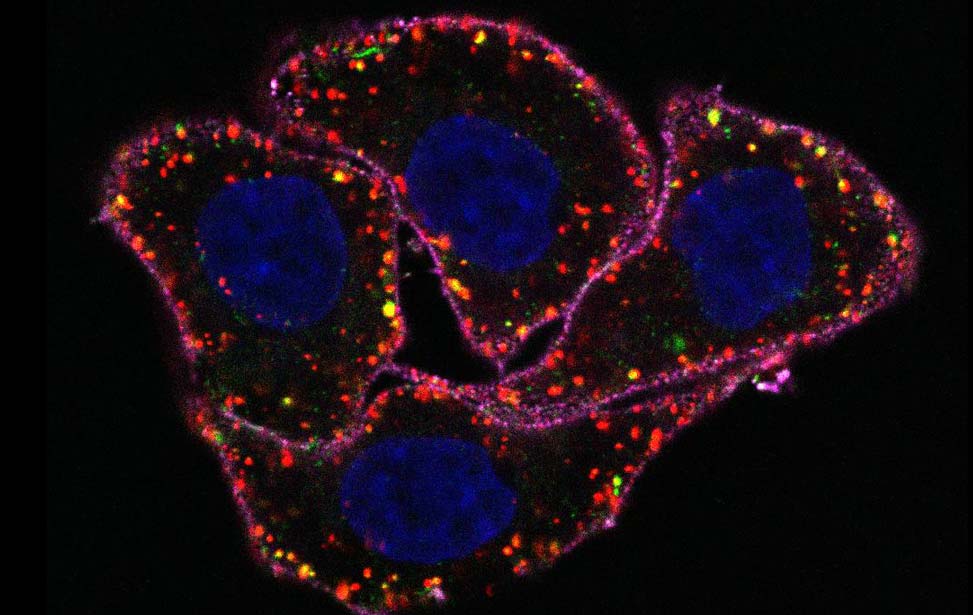
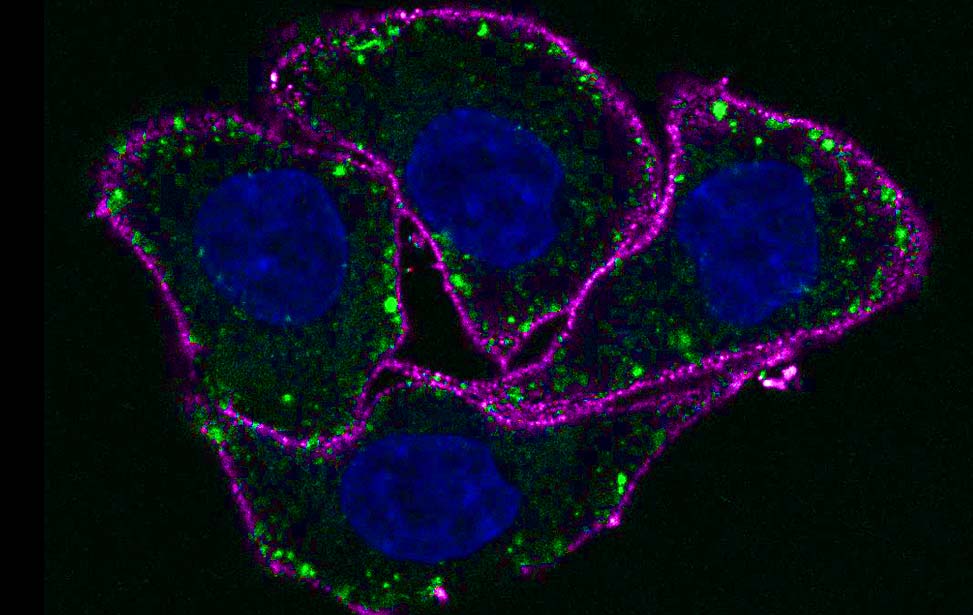
| Figure 1. The EGF-induced Ubiproteome. Upon EGF stimulation, at least 15% of the Ubiproteome was EGF-regulated (Argenzio et al, Mol Sys Biol 2011). |
In the past years, we have generated new tools and techniques that have enabled us to perform a comprehensive analysis of the impact of ubiquitin in the EGFR pathway. We have used these extensively to identify, characterize and understand specificity in the Ub binding domains recognition ( UIM, MIU and ZnFA20) and in the activity of E3 ligases (Cbl and Nedd4). Our studies will now exploit the insights we have generated (new important players and mechanisms of regulation) and the tools we have in hand.
| Figure 2. Close-up of the Ub tail in the crystal structure of HECTNedd4~UbD:Ub. When bound to the catalytic cysteine of HECTNedd4, the ubiquitin tail zips onto β-strand β9, upstream the catalytic Cys867, and organizes a hydrogen bond pattern that results in a β-sheet augmentation (Maspero et al, NSMB, 2013). |
| Figure 3. Nedd4 regulation. In resting condition, an intra-molecular interaction between the C2 and HECT domain inhibits the catalytic activity of the HECT domain. Post-translational modifications induced by RTKs (i.e. phosphorylation of specific tyr located in the interaction surface, right panel) or specific signals (i.e. increase of Ca2+ content, bottom panel) disrupt the C2:HECT interaction, generating a fully active Nedd4 (Mari, Structure, 2014; Persauld Science Signaling, 2014). |
Research project
Our laboratory is interested in understanding the role of ubiquitin in regulating fundamental processes such as endocytosis, cell migration, and autophagy, in normal and pathological conditions. To establish how ubiquitination acts, it is critical to determine the type of ubiquitin modification and to understand the mechanism by which this signal is generated. To this end, we are using a variety of biochemical, biophysical techniques, including mass spectrometry and crystallography. These techniques are coupled with classical mammalian cell-based assays and with the use of Drosophila model system (in collaboration with Thomas Vaccari in IFOM).
E3 ligases in physiology and diseasesCells sense stresses and promptly respond to them by activating a series of specific and timely-regulated processes, which allow to cope with damages and to survive. The early phase of this response precedes changes in gene expression and mainly relies on post-translational modifications, which are responsible either for the activation of the survival pathways or for the recognition of the damaged components of the cell. Enzymes that perform these modifications should therefore function as direct or indirect sensors of these stresses able to coordinate the activation of a multitude of downstream signaling pathways. Ubiquitin E3 ligases are among the best examples of such enzymes. We are investigating the function of poorly-characterized HECT E3 ligases involved in cancers and neurodegenerative diseases
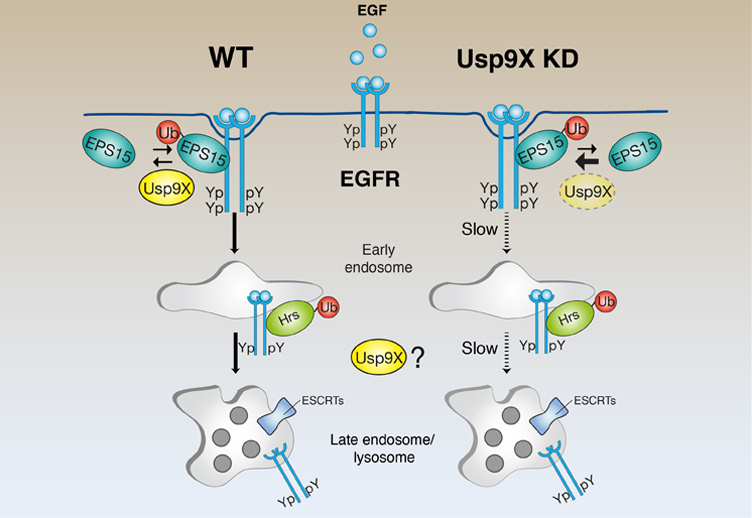
Quality control at the cell membraneIn cells, both newly synthesized and pre-existing proteins are constantly exposed to misfolding and aggregation. The accumulation of damaged proteins perturbing cellular homeostasis can cause aging, pathological states, and even cell death. Cells have developed powerful quality control strategies that counteract protein damage in a compartment-specific way. is easy to envision a "quality control" mechanism at the plasma membrane similar to the one occurring at the ER (ERAD pathway). We are investigating this possibility using the EGFR as model system.
Ubiquitin regulation in myosin VI-mediated migrationMyosin VI functions in endocytosis and cell motility and its dysfunction is associated with carcinogenesis. Alternative splicing within the myosin VI tail generates distinct isoforms but the impact of the variable exons on physiological and pathological myosin VI functions remains unknown. We recently identified a novel structured domain in myosin VI that binds to ubiquitin chains and is regulated by alternative splicing. This finding is relevant to cancer, where and exon skipping dictates cell addiction to myosin VI for tumor cell migration. We are studying this novel outcome and the mechanism behind in molecular terms.
Group members
- Elena Maspero
- Matteo Biancospino
- Carlos Niño
- Eleonora Valentini
- Janine Weber
- Valentina Fayner
- Elisa Magistrati
- Cristina Calonghi
- Michol Savio (Medical Science Manager, Bristol-Myers Squibb, Turin)
- Hans-Peter Wollscheid (Post-doc, Ulrich lab, IMB, Mainz)
- Nadine Wossner (Clinical Monitor, IZKS, Mainz)
- Sara Mari (Post-doc, Mapelli Lab, IEO, Milan)
- Wessen Maruwge (Grant Writer, IFOM, Milan)
- Filippo Acconcia (Associate Professor, Università degli Studi, Rome)
- Lorenza Penengo (Group Leader at University of Zurich)
- Elisabetta Argenzio (Post-doc, Moolenaar Lab, NKI, Amsterdam)
- Barbara Oldrini (Staff scientist, Squatrito Lab, CNIO, Madrid)
- Tanja Woelk (Post-doc, Max Planck Institute of Molecular Physiology, Dortmund)
(update: May 2016)
IFOM Review
Deubiquitination enzymes in the limelight
Commentary on Simona Polo's paper published on Current Biology, by Yossi Yarden (2015)Ubiquitin and SUMO: an hands-on experience... by the sea
Personal memories for a memorable EMBO Practical Course organized by Simona Polo, by Ronald T. Hay (2013)Illuminating a ubiquitin transfer relay, step-by-step
Commentary on Simona Polo's paper published on Nature Structural & Molecular Biology, by Brenda A. Schulman (2013)Media
In the press
Il fermo immagine della proteina multitasking
Ansa Scienza
2013.05.10 | Elisa Buson
Tumori: italiana scatta fermo immagine di proteina factotum
Ansa Salute
2013.05.10 | Elisa Buson
Una fotografia molecolare per l'ubiquitina
Scienzainrete
2013.05.10 | Francesco Aiello
Photogallery