Molecular Oncology and Immunology
Background
My group is interested in the dissection of the regulatory networks that shape human T cells functional plasticity in the context of tumor immune responses. We are mainly focused on T regulatory subsets that are central for the establishment of the tumor-dependent immunosuppressive mechanisms and ultimately contribute to dampening immune responses against cancer cells. Indeed, understanding the functional complexity and heterogeneity of human T cells cells in the tumor microenvironment is key to pave the way towards the development of novel and more specific immunomodulatory approaches for cancer treatment.
Ongoing research projects
Characterization of the molecular blueprints of T cell subsets at tumor site.
To dissect the hubs that instruct gene regulatory programs of immune cell landscape at tumor sites, we employ whole transcriptome along with single-cell RNA-sequencing that provides a thorough description of the underlying heterogeneity of T cell subsets in the tumor and metastatic microenvironment. Moreover, we take advantage of bioinformatic and computational pipelines to integrate transcriptome datasets with epigenomic profiling and unveil also cis-regulatory elements (e.g. enhancers) and other non-coding genomic features (e.g. long non-coding RNAs) that shape the identity of tumor infiltrating lymphocytes.
Charting the communication routes between immune and tumor cells.
A key aspect of tumor immune response is the crosstalk between tumor and immune cells, whose function is strongly influenced by their spatial localization and tissue context. To chart cell-spatial organization at tumor sites we are employing both enhanced single-molecule FISH approaches on pre-selected targets and spatial transcriptomics techniques based on in situ mRNA capturing coupled to standard RNA-seq (SLIDE-seq).
Modelling tumor-immune host interactions through organoid 3D cultures.
To fully understand the functional implication of the relationships between immune and tumor cells we are also exploiting organoids cell lines derived from both tumor and normal tissues. Organoids recapitulate the architecture and growth pattern of the tissue from which they originate and represent therefore an experimentally controllable model to assess the modulation of either tumor or T cell specific transcripts by gene editing approaches in an environment resembling the original tumor.
Phd project title
- Development of bioinformatics methods for the integration of transcriptomic and epigenomic analyses of colorectal cancer derived organoids
- Epigenetic characterization of tumor infiltrating CD4+ T regulatory cells for identification of novel anti-cancer targets
- Functional characterization of long non-coding RNAs that shape regulatory T cell identity
29 june 2020 rel.02
Photogallery
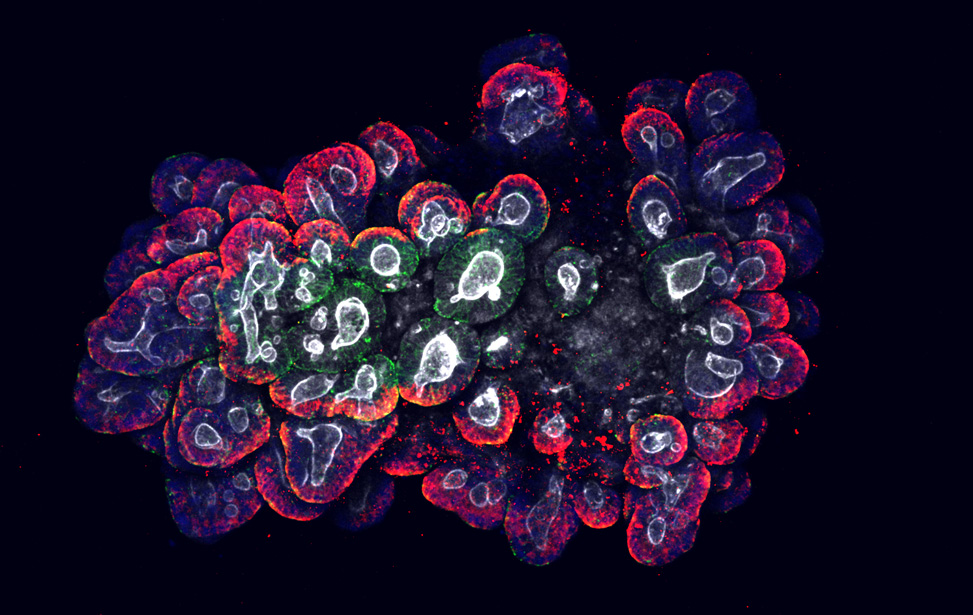 Maximum projection of human colorectal cancer patients derived organoids (CRC PDO)
Maximum projection of human colorectal cancer patients derived organoids (CRC PDO)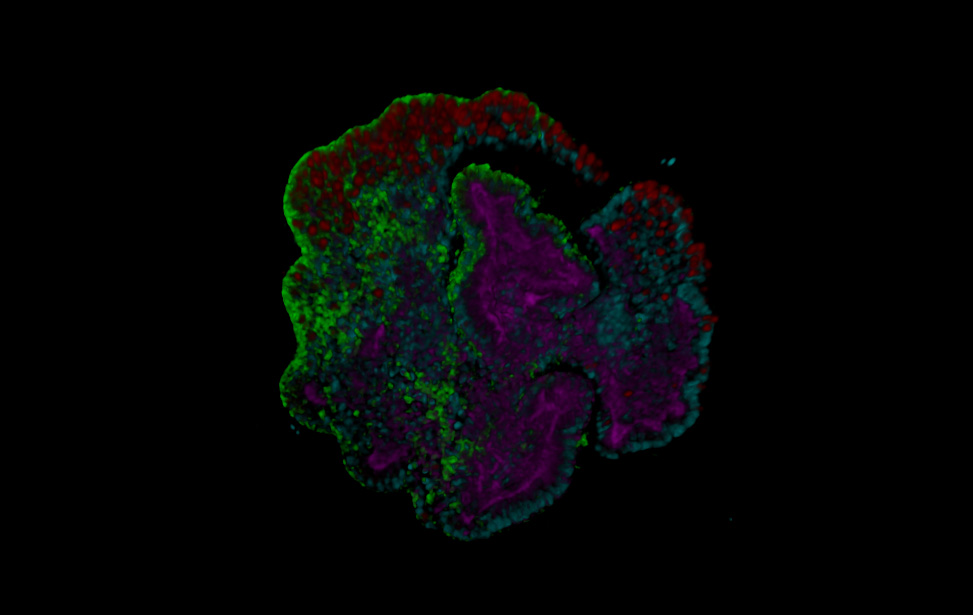 3D rendering of human CRC PDO
3D rendering of human CRC PDO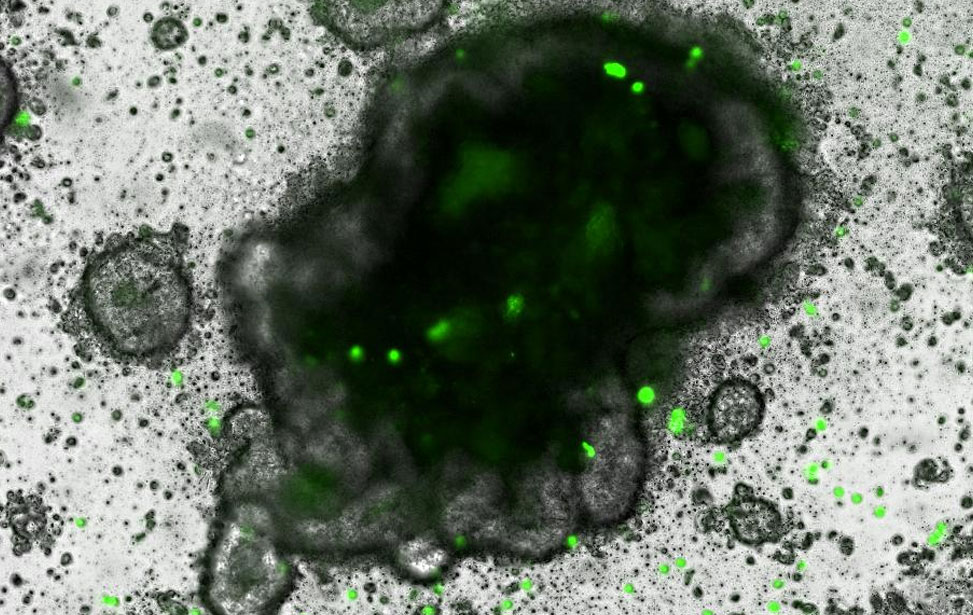 Co-culture of autologous human tumor infilitrating lymphocytes and CRC PDO
Co-culture of autologous human tumor infilitrating lymphocytes and CRC PDO 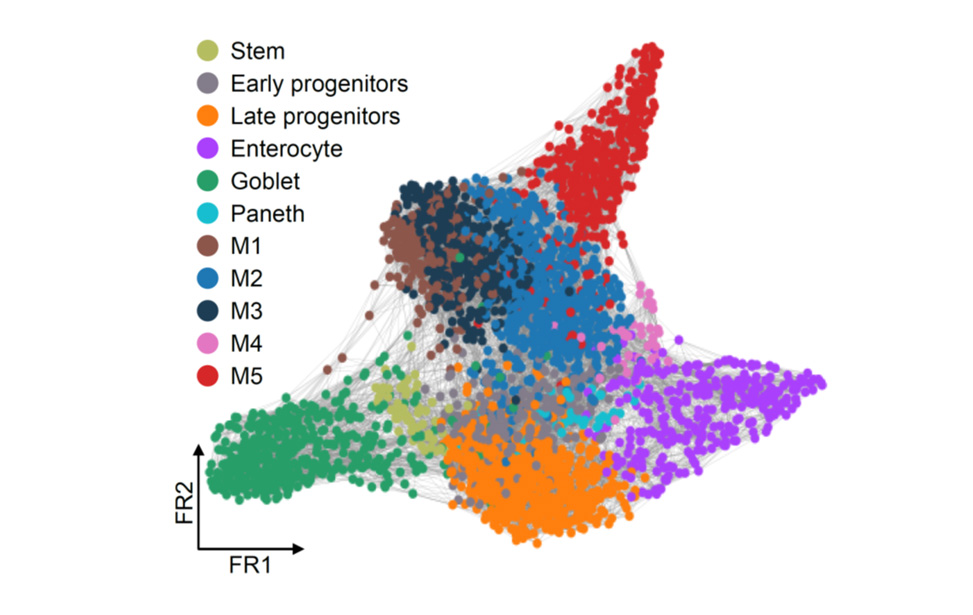 scRNA seq analysis of human CRC tissue
scRNA seq analysis of human CRC tissue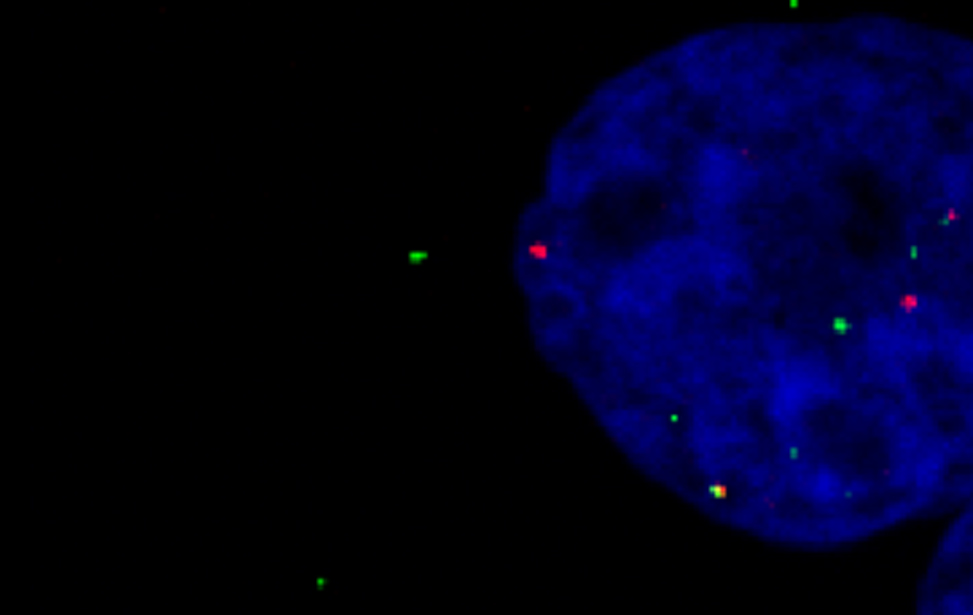 smRNA FISH in human primary T lymphocytes
smRNA FISH in human primary T lymphocytes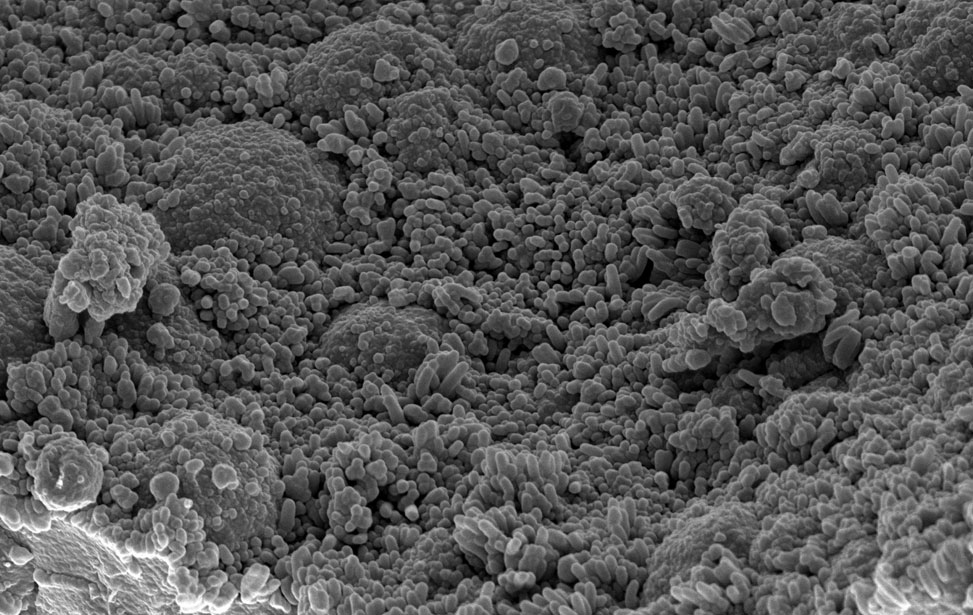 Transmission electron microscopy (TEM) of human CRC PDO lumen
Transmission electron microscopy (TEM) of human CRC PDO lumen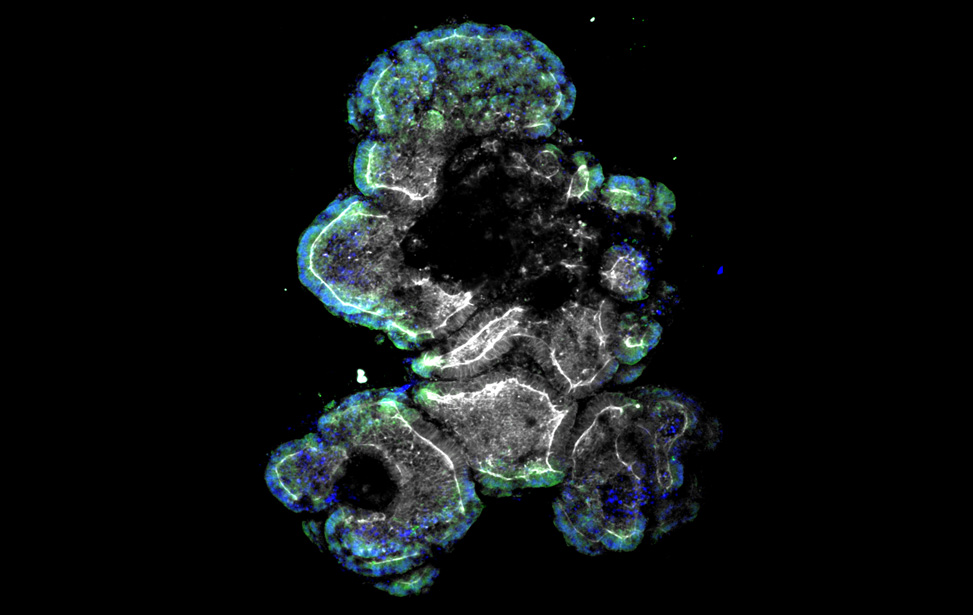 Human colon liver metastasis patients derived organoids
Human colon liver metastasis patients derived organoids